Equilibrium sampling approach to the interpretation of electron density maps
| Title | Equilibrium sampling approach to the interpretation of electron density maps |
| Publication Type | Journal Article |
| Year of Publication | 2014 |
| Authors | Vitalis A., Caflisch A. |
| Journal | Structure |
| Volume | 22 |
| Issue | 1 |
| Pagination | 156-167 |
| Date Published | 2014 Jan 7 |
| Type of Article | Research Article |
| Keywords | Actin-Related Protein 2-3 Complex, Computer Simulation, Crystallography, X-Ray, Electrons, Humans, Inverted Repeat Sequences, Kinetics, Models, Molecular, RNA, Thermodynamics, Ubiquitin |
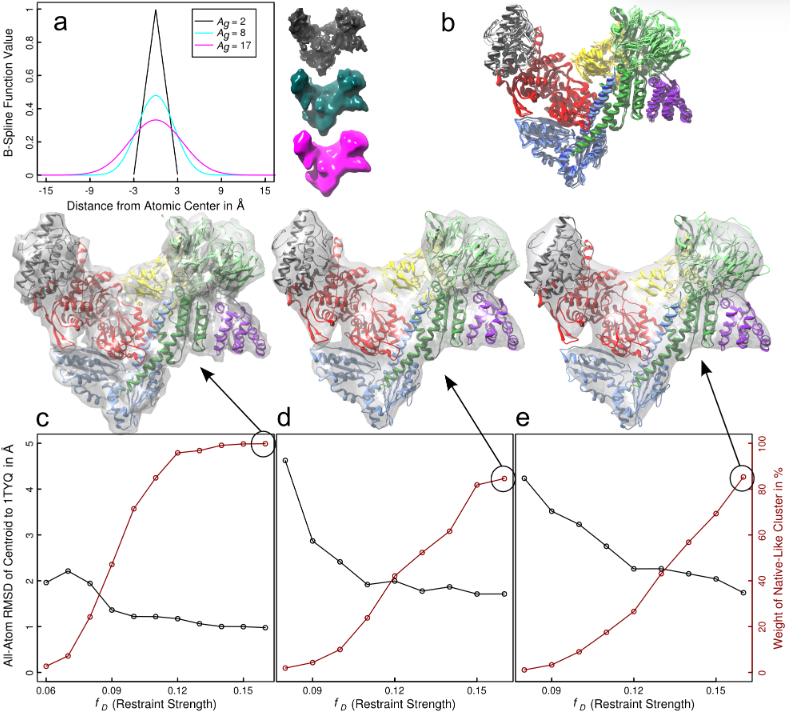
| Abstract | The derivation of molecular models from spatial density data generated by X-ray crystallography or electron microscopy is an active field of research. Here, we introduce and evaluate an approach relying on the equilibrium sampling of energy landscapes describing restraints to experimental input data. Our procedure combines density restraints with replica exchange methodologies in the parameter space of the restraints, and we demonstrate its applicability to both flexible polymers and the assembly of protein complexes from rigid components. For the most difficult system studied, we highlight the importance of advanced data analysis techniques in mining poorly converged data further. Successful and unbiased interpretation of input density maps is a prerequisite for using this approach as an auxiliary restraint term in molecular simulations. Because these simulations will also utilize physical interaction potentials, we hope that they will contribute to deriving families of structural models for input data that are ambiguous per se. |
| DOI | 10.1016/j.str.2013.10.014 |
| pubindex | 0178 |
| Alternate Journal | Structure |
| PubMed ID | 24316403 |